Ribonucleotide monophosphates (rNMPs) and deoxyribonucleotide monophosphates are the basic building blocks of RNA and DNA. The difference between them is that rNMPs contain ribose instead of deoxyribose as their sugar component. During processes like DNA replication and repair, rNMPs become embedded in genomic DNA, influencing DNA fragility, mutability, and ultimately the stability of the genome.
Because rNMPs alter the way DNA works in both structure and function, it’s important to be able to identify them and their sites of genomic incorporation. Recent advances in high-throughput sequencing techniques now make it possible to tag rNMPs embedded in genomic DNA. Simultaneously with three other methods to capture rNMPs in DNA, a unique and robust technique called ribose-seq was developed by the lab of Francesca Storici, professor in the School of Biological Sciences at the Georgia Institute of Technology, and her collaborators.
They published a description of the technique and the discoveries it yielded a few years ago in the journal Nature Methods. While their method is applicable to DNA from virtually any source and organism (including humans), allowing the researchers to determine the full profile of rNMPs embedded in genomic DNA, it essentially generates large, complex datasets like the other three approaches to study ribonucleotides in DNA.
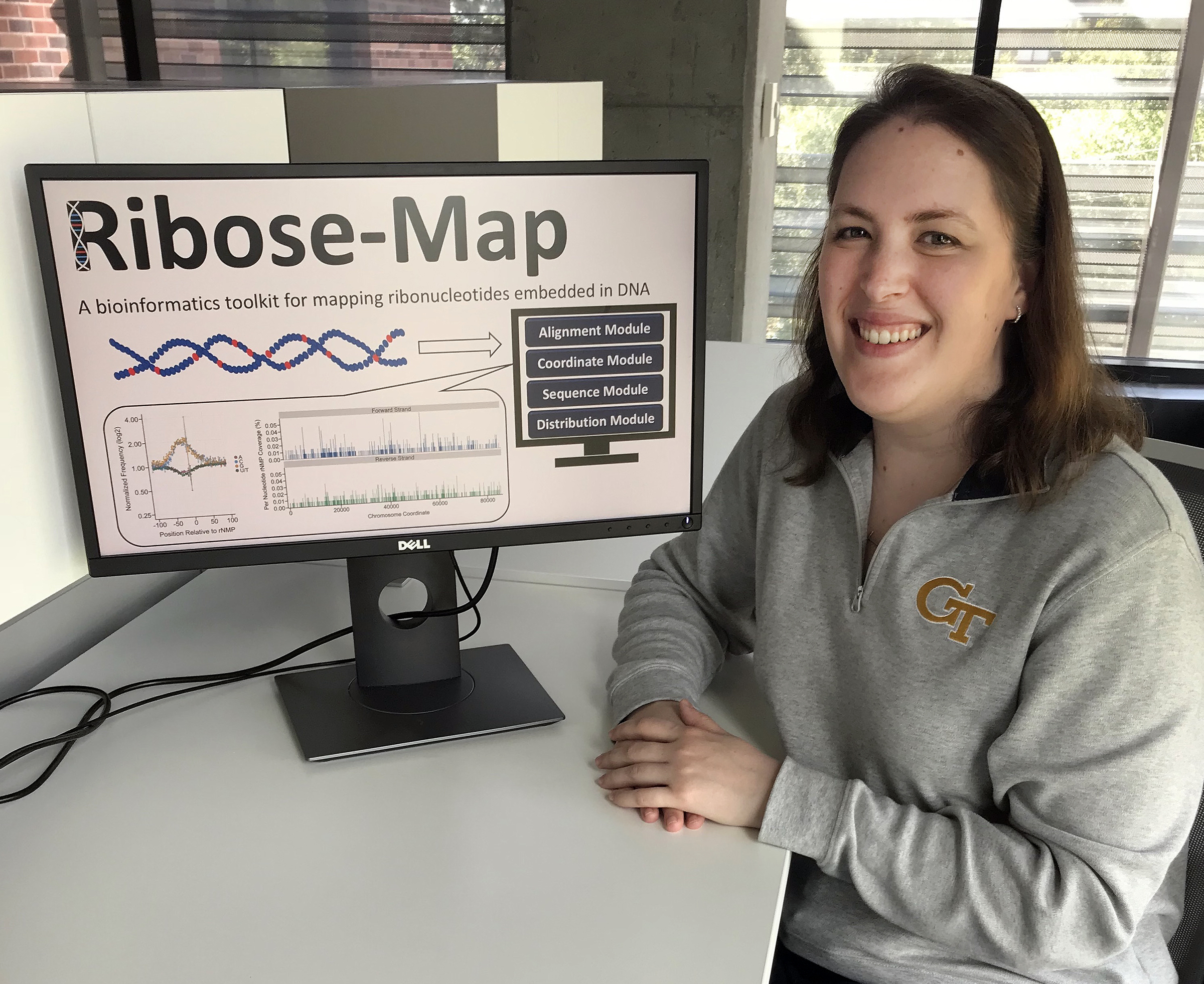
“But there is no standardized system to analyzing the data from ribose-seq or the three other techniques,” notes Alli Gombolay, a fourth-year PhD student in Storici’s lab. “We wanted to create a bioinformatics toolkit that could rapidly and effectively analyze the data from any of those techniques to study all types of data and gather as much information as possible.”
Standard computational pipelines designed to map embedded rNMPs are customized for data generated using only one kind of sequencing technique. So Gombolay and her co-advisors – Storici and Fred Vannberg, researchers in the Petit Institute for Bioengineering and Bioscience – developed Ribose-Map.
They recently published their research in the journal Nucleic Acids Research. In their paper, entitled “Ribose-Map: a bioinformatics toolkit to map ribonucleotides embedded in DNA,” they describe how to transform raw sequencing data into summary datasets and publication-ready results, which would allow researchers to identify sites of embedded rNMPs, study the nucleotide sequence context of these rNMPs, and explore their genome-wide distribution.
“Ribose-Map increases reproducibility and allows us to directly compare the data, and ultimately gather more information.” says Gombolay.
Other labs are already interested in making those comparisons. This became abundantly clear to Gombolay in September when she attended the 15th RNase H meeting in Warsaw, Poland, a biennial international gathering.
For researchers who want to know more about Ribose-Map and how to use it, Gombolay has created a GitHub page, where she describes how to set-up, install, and run the toolkit.
“It’s a simpler approach to mapping ribose in DNA or other modifications,” she says. “It’s particularly helpful for people with limited bioinformatics skillsets, or for people who are new to the relatively small field of ribonucleotide mapping. But since we can now map ribonucleotides in DNA, we think the field could grow faster.”
For More Information Contact
Jerry Grillo
Communications Officer II
Parker H. Petit Institute for
Bioengineering and Bioscience